Vitiligo: NLRP1
Phylogeny

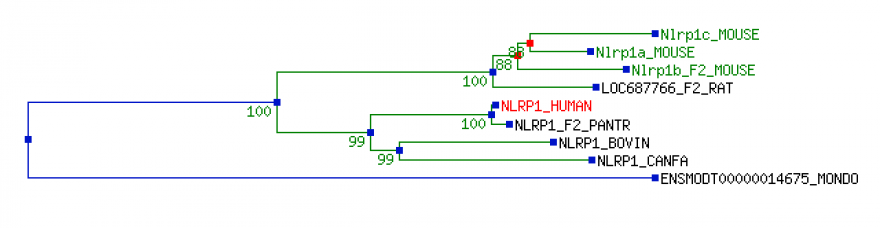
The tree shown below was obtained from Tree Fam [1], a database of phylogenetic trees of animal genes. The tree is a clean tree, which means that it only contains species that have been sequenced. Clean trees are built by merging several trees together, including Phyml WAG tree, Phyml HKY, NJ dS and NJ dN trees. Branch lengths are estimated using the HKY model. This information was obtained from Tree Fam [1].
Humans share most of their traits with chimpanzees. This is evidenced by the small evolutionary distances between their diverging branches. Additionally, this tree shows that the human NLRP1 gene shows homology with three different paralogs of the mouse Nlrp1 genes.
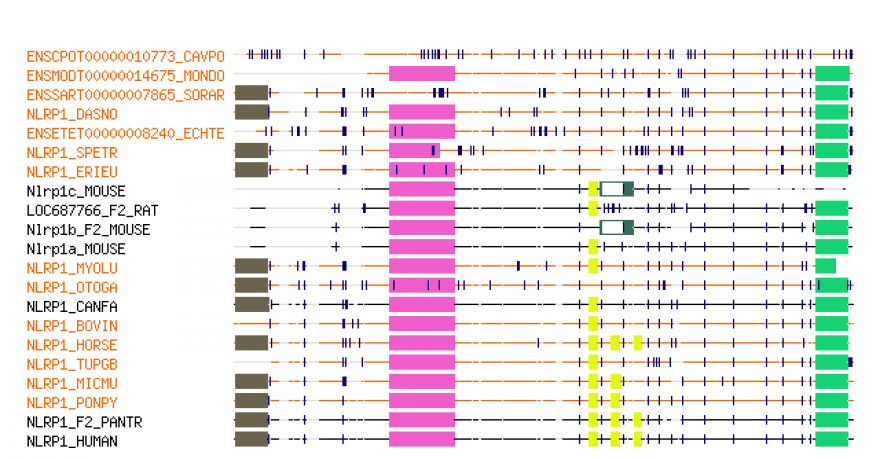
Below is a visualization of an alignment obtained from Tree Fam [1]. Boxes correspond to Pfam domains, while lines correspond to non-domain regions. Dark lines or boxes show matched parts, while light ones show gaps. Vertical dark blue lines indicate splicing boundaries.

Phylogeny.fr [2]
Construction parameters:
Multiple Alignment: Clustal W
Alignment Curation: GBlocks
Construction: TNT (Parsimony)
Visualisation: TreeDyn
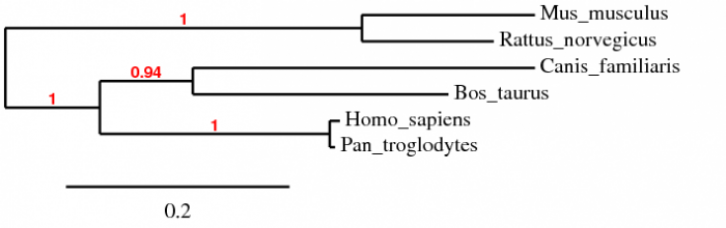
Multiple Alignment: Clustal W
Alignment Curation: GBlocks
Construction: TNT (Parsimony)
Visualisation: TreeDyn
| phylo_tree-1_protein_parsimony.png |
| homologeneprotein.txt |
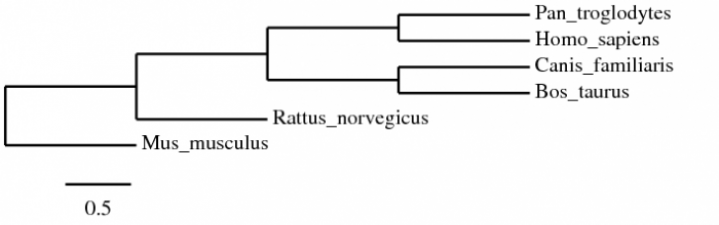
Multiple Alignment: MUSCLE
Alignment Curation: GBlocks
Construction: PhyML (Maximum Likelihood)
Visualisation: TreeDyn
| phylo_tree_protein_ml.png |
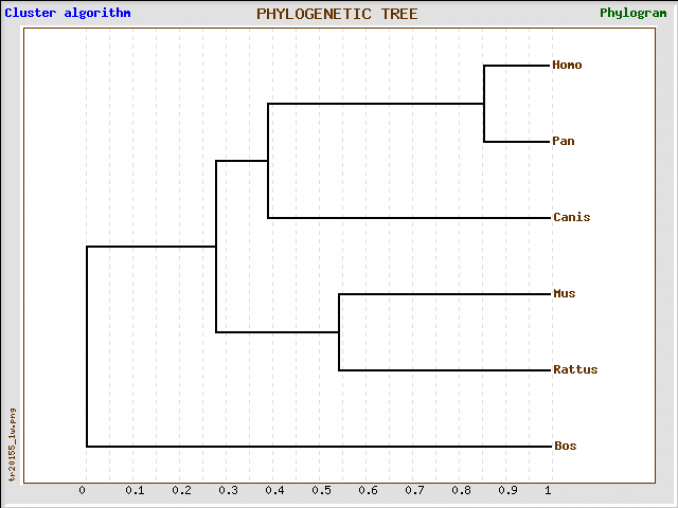
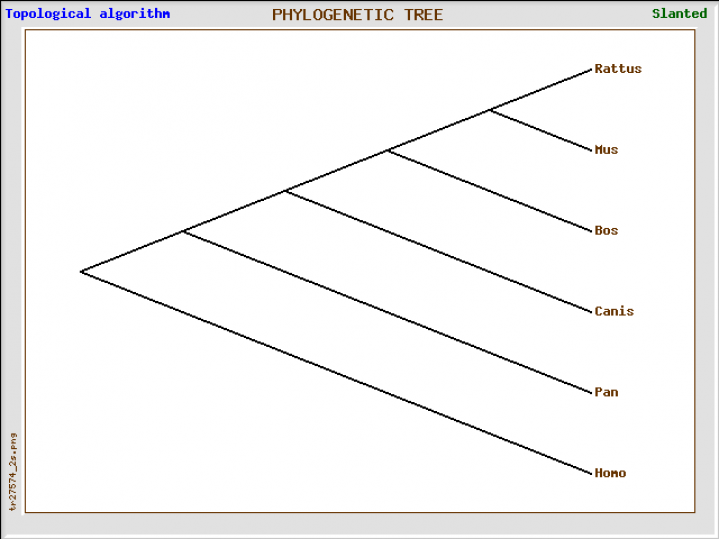
This tree was generated by inputting FASTA protein sequence into phylogenetic program TreeTop [4]. A cluster algorithm and a topological algorithm were both used to generate trees.
NACHT Domain Tree
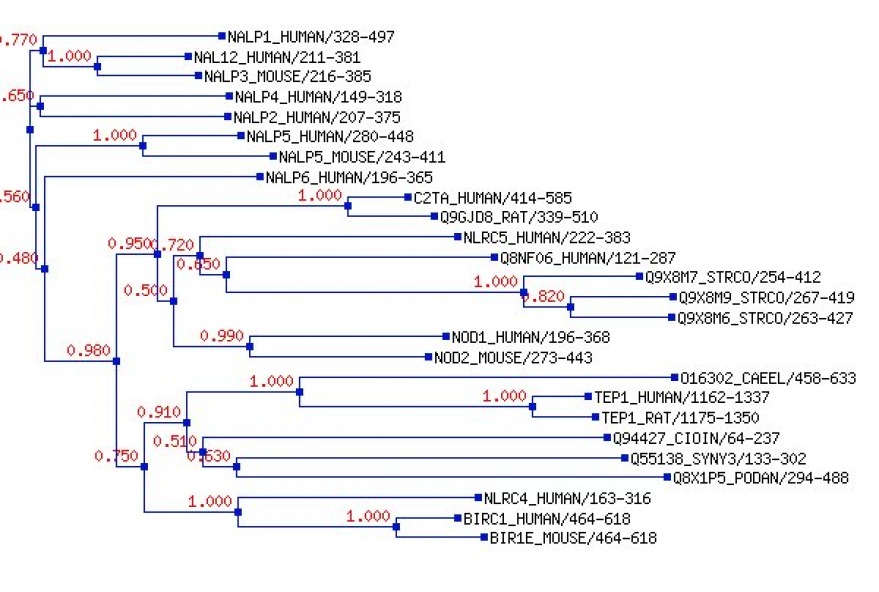
NACHT domain tree generated by Fast Tree via Pfam.
Analysis
While different parameters were used to generate phylogenetic trees for the NLRP1 protein sequence and its respective protein domains, the results yielded are generally the same. Humans share their most recent history with the P. troglodytes, otherwise known as chimpanzees. From their, they show strong phylogenetic relationships with cow and dog species. The Mus musculus (mouse) has three different paralogs that correspond to the human NLRP1 gene and protein.
This Web page was created as an assignment for Genetics 677 at UW-Madison, Spring 2010.
Sarah Hamilton, May 16, 2010.
Sarah Hamilton, May 16, 2010.