Vitiligo: NLRP1
Motifs

Using motif-detecting online software programs, such as MEME [1], short, recurring patterns of DNA that are presumed to have biological function were selected for. Often, these motifs indicate sequence-specific binding sites for proteins or are involved in important processes at the RNA level, including ribosome binding, mRNA splicing, and transcription termination.
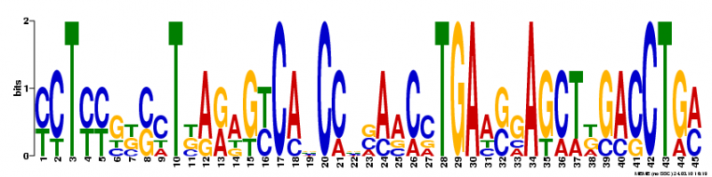
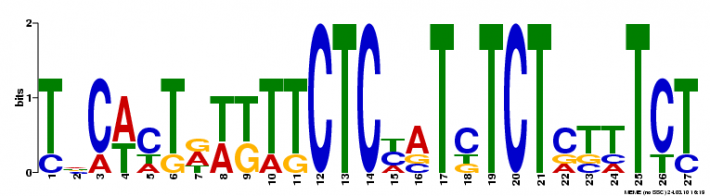
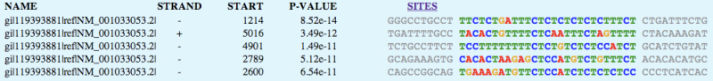
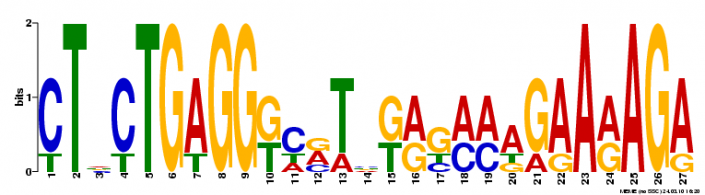
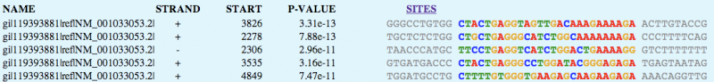
NLRP1 FASTA-formatted mRNA sequence was used to detect DNA motifs in the NLRP1 gene. mRNA from NLRP1 isoform 5 was used. Results were generated using a motif-detecting online software program called MEME [1].
Motifs were searched for using the following parameters:
Distribution of motifs among sequences: anywhere
Minimum width: 6
Maximum width: 50
Maximum number of motifs to find: 25
MEME generated 25 motifs using these parameters. Three of these motif findings are shown below.
| meme.txt |

Motif Finder [2]
NLRP1 FASTA-formatted mRNA sequence was also used in a motif-detecting software program called MOTIF [2].
Results:
Cutoff Motifs Found
85(default) 11
95 11
100 11
Motif Descriptions
1. EGF-like domain signature 1
2. Insulin-like growth factor binding proteins signature
3. Integrins beta chain cysteine-rich domain signature
4. C-terminal cystine knot signature
5. Anaphylatoxin domain signature
6. 4Fe-4S ferredoxins, iron-sulfur binding region signature
7. Thiolases active site
8. Tubulin subunits alpha, beta, and gamma signature
9. VWFC domain signature
10. 2 Fe-2S ferredoxins, iron-sulfur binding region signature
11. Mammalian defensins signature
Analysis
By inputting a FASTA-formatted mRNA sequence of NLRP1 isoform 5, MOTIF generated results using varied parameters. I began my search with a cut-off score of 85 - the default setting. This generated 11 results for motifs. I increased the stringency of my motif search by increasing the cut-off score from 85 to 95 to 100. Each search I conducted yielded 11 motifs. The results of my query were returned almost instantly. However, detailed information of the motifs (e.g. function of motif) was lacking.
Results:
Cutoff Motifs Found
85(default) 11
95 11
100 11
Motif Descriptions
1. EGF-like domain signature 1
2. Insulin-like growth factor binding proteins signature
3. Integrins beta chain cysteine-rich domain signature
4. C-terminal cystine knot signature
5. Anaphylatoxin domain signature
6. 4Fe-4S ferredoxins, iron-sulfur binding region signature
7. Thiolases active site
8. Tubulin subunits alpha, beta, and gamma signature
9. VWFC domain signature
10. 2 Fe-2S ferredoxins, iron-sulfur binding region signature
11. Mammalian defensins signature
Analysis
By inputting a FASTA-formatted mRNA sequence of NLRP1 isoform 5, MOTIF generated results using varied parameters. I began my search with a cut-off score of 85 - the default setting. This generated 11 results for motifs. I increased the stringency of my motif search by increasing the cut-off score from 85 to 95 to 100. Each search I conducted yielded 11 motifs. The results of my query were returned almost instantly. However, detailed information of the motifs (e.g. function of motif) was lacking.
References
[1] Timothy L. Bailey and Charles Elkan, "Fitting a mixture model by expectation maximization to discover motifs in biopolymers", Proceedings of the Second International Conference on Intelligent Systems for Molecular Biology, pp. 28-36, AAAI Press, Menlo Park, California, 1994.
[2] Motif: http://motif.genome.jp/
[2] Motif: http://motif.genome.jp/
This Web site was created as a project for Genetics 677 at UW-Madison, Spring 2010.
May 16, 2010. Sarah Hamilton.
May 16, 2010. Sarah Hamilton.